publications
You can also find my articles on my Google Scholar.
2026
- ICLR 2026
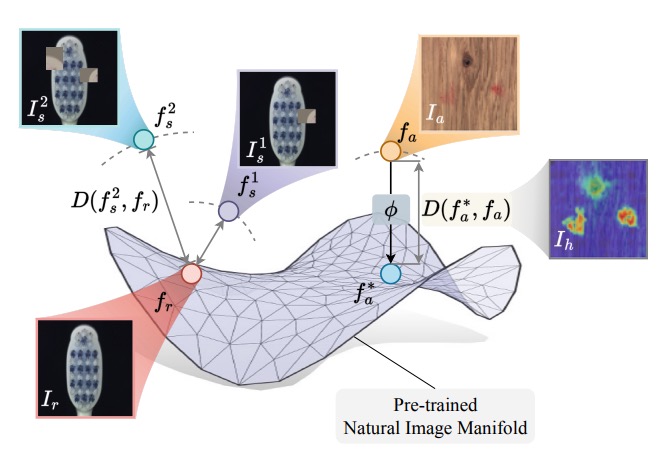 Foundation Visual Encoders Are Secretly Few-Shot Anomaly DetectorsGuangyao Zhai* , Yue Zhou*, Xinyan Deng, and 3 more authorsInternational Conference on Learning Representations, 2026* equal contribution
Foundation Visual Encoders Are Secretly Few-Shot Anomaly DetectorsGuangyao Zhai* , Yue Zhou*, Xinyan Deng, and 3 more authorsInternational Conference on Learning Representations, 2026* equal contributionFew-shot anomaly detection streamlines and simplifies industrial safety inspection. However, limited samples make accurate differentiation between normal and abnormal features challenging, and even more so under category-agnostic conditions. Large-scale pre-training of foundation visual encoders has advanced many fields, as the enormous quantity of data helps to learn the general distribution of normal images. We observe that the anomaly amount in an image directly correlates with the difference in the learnt embeddings and utilize this to design a few-shot anomaly detector termed FoundAD. This is done by learning a nonlinear projection operator onto the natural image manifold. The simple operator acts as an effective tool for anomaly detection to characterize and identify out-of-distribution regions in an image. Extensive experiments show that our approach supports multi-class detection and achieves competitive performance while using substantially fewer parameters than prior methods. Backed up by evaluations with multiple foundation encoders, including fresh DINOv3, we believe this idea broadens the perspective on foundation features and advances the field of few-shot anomaly detection.
- AAAI 2026
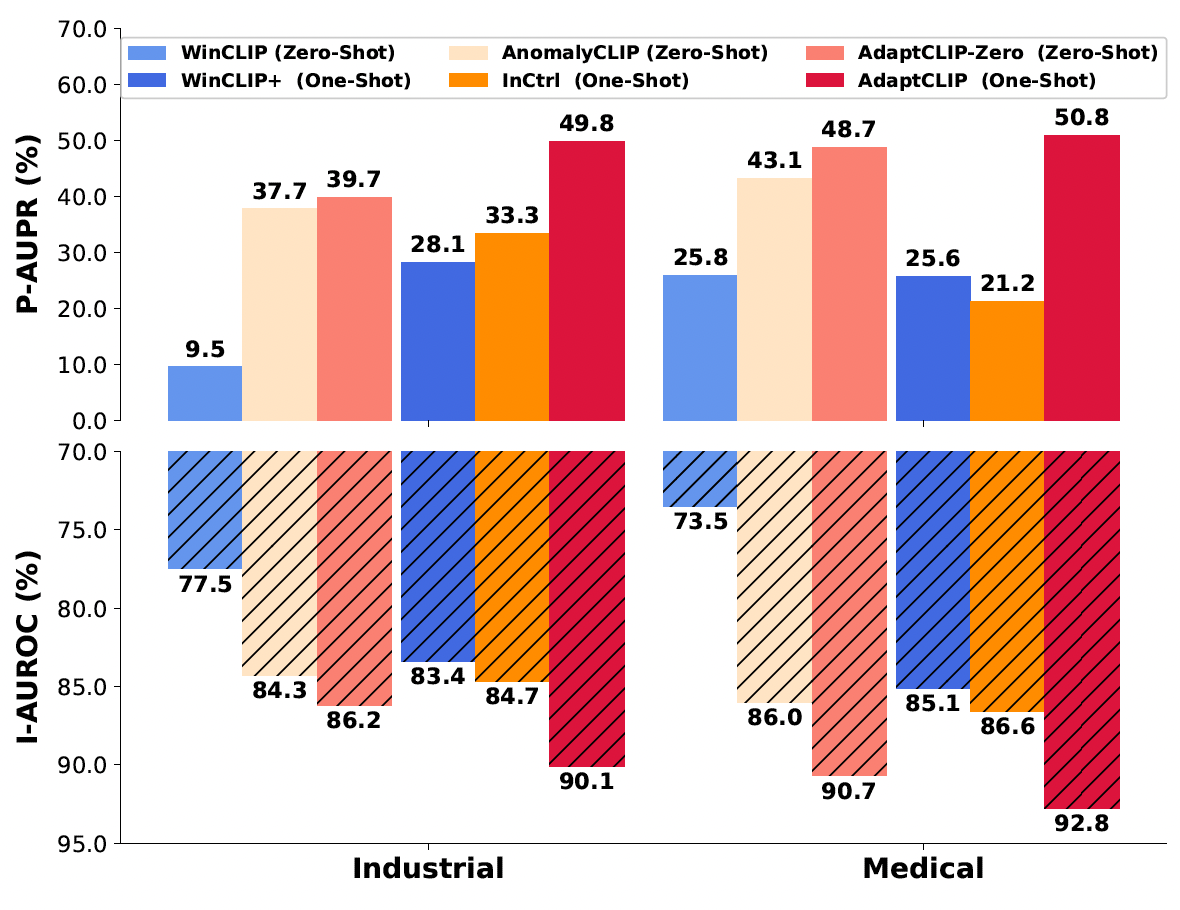 AdaptCLIP: Adapting CLIP for Universal Visual Anomaly DetectionBin-Bin Gao , Yue Zhou, Jiangtao Yan, and 7 more authorsAnnual AAAI Conference on Artificial Intelligence, 2026
AdaptCLIP: Adapting CLIP for Universal Visual Anomaly DetectionBin-Bin Gao , Yue Zhou, Jiangtao Yan, and 7 more authorsAnnual AAAI Conference on Artificial Intelligence, 2026Universal visual anomaly detection aims to identify anomalies from novel or unseen vision domains without additional fine-tuning, which is critical in open scenarios. Recent studies have demonstrated that pre-trained vision-language models like CLIP exhibit strong generalization with just zero or a few normal images. However, existing methods struggle with designing prompt templates, complex token interactions, or requiring additional fine-tuning, resulting in limited flexibility. In this work, we present a simple yet effective method called AdaptCLIP based on two key insights. First, adaptive visual and textual representations should be learned alternately rather than jointly. Second, comparative learning between query and normal image prompt should incorporate both contextual and aligned residual features, rather than relying solely on residual features. AdaptCLIP treats CLIP models as a foundational service, adding only three simple adapters, visual adapter, textual adapter, and prompt-query adapter, at its input or output ends. AdaptCLIP supports zero-/few-shot generalization across domains and possesses a training-free manner on target domains once trained on a base dataset. AdaptCLIP achieves state-of-the-art performance on 12 anomaly detection benchmarks from industrial and medical domains, significantly outperforming existing competitive methods.
2025
- MICCAI 2025
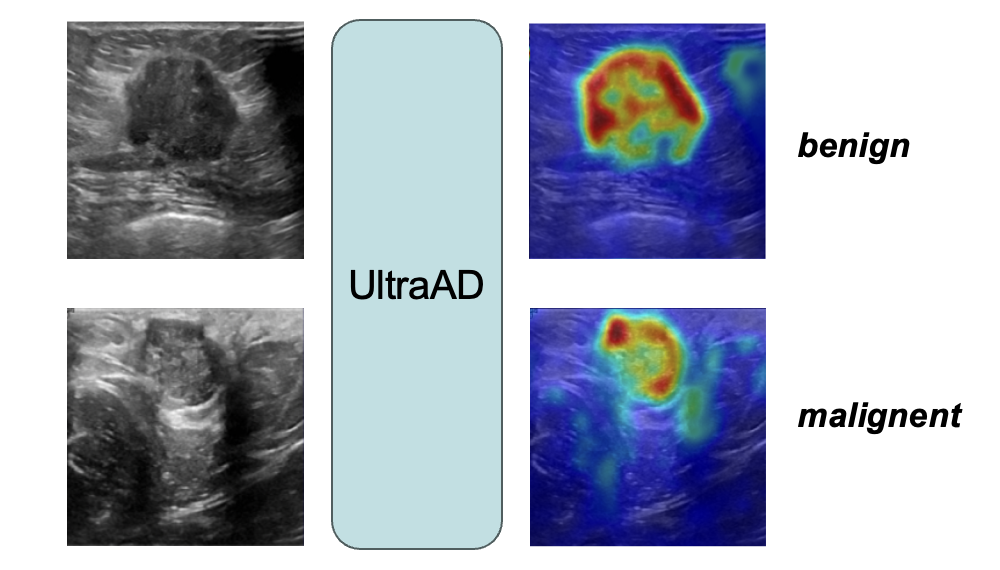 UltraAD: Fine-Grained Ultrasound Anomaly Classification via Few-Shot CLIP AdaptationYue Zhou, Yuan Bi, Wenjuan Tong, and 3 more authorsMedical Image Computing and Computer Assisted Intervention , 2025
UltraAD: Fine-Grained Ultrasound Anomaly Classification via Few-Shot CLIP AdaptationYue Zhou, Yuan Bi, Wenjuan Tong, and 3 more authorsMedical Image Computing and Computer Assisted Intervention , 2025Precise anomaly detection in medical images is critical for clinical decision-making. While recent unsupervised or semi-supervised anomaly detection methods trained on large-scale normal data show promising results, they lack fine-grained differentiation, such as benign vs. malignant tumors. Additionally, ultrasound (US) imaging is highly sensitive to devices and acquisition parameter variations, creating significant domain gaps in the resulting US images. To address these challenges, we propose UltraAD, a vision-language model (VLM)-based approach that leverages few-shot US examples for generalized anomaly localization and fine-grained classification. To enhance localization performance, the image-level token of query visual prototypes is first fused with learnable text embeddings. This image-informed prompt feature is then further integrated with patch-level tokens, refining local representations for improved accuracy. For fine-grained classification, a memory bank is constructed from few-shot image samples and corresponding text descriptions that capture anatomical and abnormality-specific features. During training, the stored text embeddings remain frozen, while image features are adapted to better align with medical data. UltraAD has been extensively evaluated on three breast US datasets, outperforming stateof-the-art methods in both lesion localization and fine-grained medical classification. The code will be released upon acceptance.
@misc{zhou2025ultraadfinegrainedultrasoundanomaly, title = {UltraAD: Fine-Grained Ultrasound Anomaly Classification via Few-Shot CLIP Adaptation}, author = {Zhou, Yue and Bi, Yuan and Tong, Wenjuan and Wang, Wei and Navab, Nassir and Jiang, Zhongliang}, journal = {Medical Image Computing and Computer Assisted Intervention}, year = {2025}, }
2023
- MIA 2023
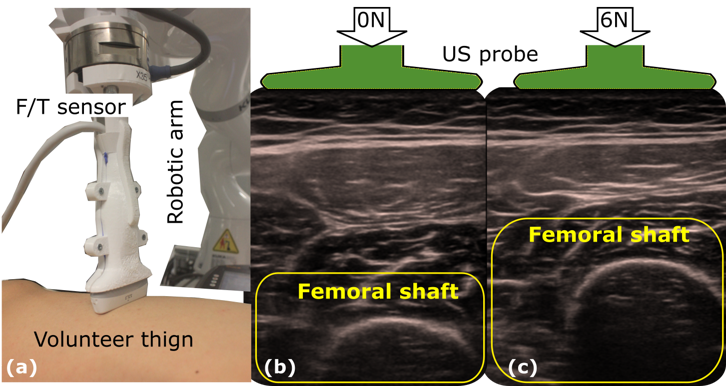 DefCor-Net: Physics-aware ultrasound deformation correctionMedical Image Analysis, 2023* equal contribution
DefCor-Net: Physics-aware ultrasound deformation correctionMedical Image Analysis, 2023* equal contributionThe recovery of morphologically accurate anatomical images from deformed ones is challenging in ultrasound (US) image acquisition, but crucial to accurate and consistent diagnosis, particularly in the emerging field of computer-assisted diagnosis. This article presents a novel anatomy-aware deformation correction approach based on a coarse-to-fine, multi-scale deep neural network (DefCor-Net). To achieve pixel-wise performance, DefCor-Net incorporates biomedical knowledge by estimating pixel-wise stiffness online using a U-shaped feature extractor. The deformation field is then computed using polynomial regression by integrating the measured force applied by the US probe. Based on real-time estimation of pixel-by-pixel tissue properties, the learning-based approach enables the potential for anatomy-aware deformation correction. To demonstrate the effectiveness of the proposed DefCor-Net, images recorded at multiple locations on forearms and upper arms of six volunteers are used to train and validate DefCor-Net. The results demonstrate that DefCor-Net can significantly improve the accuracy of deformation correction to recover the original geometry (Dice Coefficient: from 14.3±20.9 to 82.6±12.1 when the force is 6N).
@article{jiang2023defcor, author = {Jiang, Zhongliang and Zhou, Yue and Cao, Dongliang and Navab, Nassir}, title = {DefCor-Net: Physics-aware ultrasound deformation correction}, journal = {Medical Image Analysis}, year = {2023}, note = {* equal contribution} }
2021
- RAL 2021
 Deformation-Aware Robotic 3D UltrasoundIEEE Robotics and Automation Letters(Oral presentation in IROS 2021) , 2021* equal contribution
Deformation-Aware Robotic 3D UltrasoundIEEE Robotics and Automation Letters(Oral presentation in IROS 2021) , 2021* equal contributionTissue deformation in ultrasound (US) imaging leads to geometrical errors when measuring tissues due to the pressure exerted by probes. Such deformation has an even larger effect on 3D US volumes as the correct compounding is limited by the inconsistent location and geometry. This work proposes a patient- specified stiffness-based method to correct the tissue deformations in robotic 3D US acquisitions. To obtain the patient-specified model, robotic palpation is performed at sampling positions on the tissue. The contact force, US images and the probe poses of the palpation procedure are recorded. The contact force and the probe poses are used to estimate the nonlinear tissue stiffness. The images are fed to an optical flow algorithm to compute the pixel displacement. Then the pixel-wise tissue deformation under different forces is characterized by a coupled quadratic regression. To correct the deformation at unseen positions on the trajectory for building 3D volumes, an interpolation is performed based on the stiffness values computed at the sampling positions. With the stiffness and recorded force, the tissue displacement could be corrected. The method was validated on two blood vessel phantoms with different stiffness. The results demonstrate that the method can effectively correct the force-induced deformation and finally generate 3D tissue geometries.